Model Systems of CRC and IBD for “treatable” biomarker identification
The complex nature of CRC integrating genetic, epigenetic, environmental and microbial cues underscores the need for a holistic perspective and suggests that assessing these factors combinatorially on a personalized basis may be the key to pinpoint them. Moreover, CRC studies necessitate the use of simple model hosts for causality assessment. Model hosts can reduce the complexity of the disease while reflecting key aspects of the human histopathology and concomitant molecular signals. Mice and fruit flies possess these two key properties and are thus widely used. We have developed a Drosophila stem cell tumororigenesis model and a tumor cell migration model and corresponding biomarkers, which are “treatable” depending on diet and intestinal microbes.
Based on data from human, mouse and Drosophila studies, we point to the importance of interactions among host gene expression, the intestinal microbiome and environment and systemic factors and metabolites, which comprise the intestinal holo’ome, an integral system controlling homeostasis, inflammation and cancer. As a roadmap for future studies on intestinal holo’omes we propose: a) a synthesis of information on individual human genome/epigenome, which ultimately shape the transcriptome, proteome and metabolome, the microbiota metagenome that shapes microbial transcriptome and proteome, the fecal metabolome and proteome and the blood secretome at critical time points, long before and upon the development of pre-cancerous lesions; b) the identification of combinatorial factors as potential detrimental synergisms within holo’omes linked to disease onset; c) the validation of such synergisms using fly and mouse models; and d) the assessment of therapeutics against such detrimental synergisms in clinical trials.
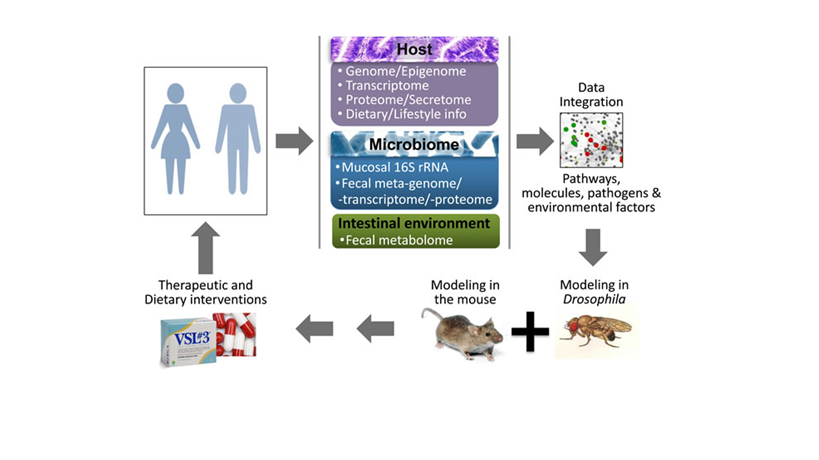
Figure: A roadmap to identify detrimental synergisms within human holo’omes as causal for colon cancer and develop personalized therapeutic or preventive strategies. A systems biology approach to assess shifts in intestinal holo’omes in humans and its link to colorectal pathologies will necessitate analysis of host intestine and microbial community genome, transcriptome, proteome metabolome and blood secretome. Using computational platforms, the genetic, metabolic, nutritional, microbial and immunological information accumulated, together with publicly available phenotypic and molecular function data, will be explored to obtain a ‘holistic’ view of key pathogenic processes and their hierarchies, to simulate the expected response to hypothetical interventions and develop new basic and translational research hypotheses. Reductionist approaches in Drosophila and mice – which can be genetically manipulated to express or lose the expression of specific genes in the intestine, while fed or injected with specific microbes and metabolites – could be used to assess detrimental synergisms of the intestinal holo’ome in driving inflammation and tumorigenesis, and guide the development of intervention strategies. Such therapeutic or dietary interventions could be translated to the clinic aiming to treat patients against microbial and intestinal environment imbalances as a means to alleviate intestinal inflammation and CRC.
