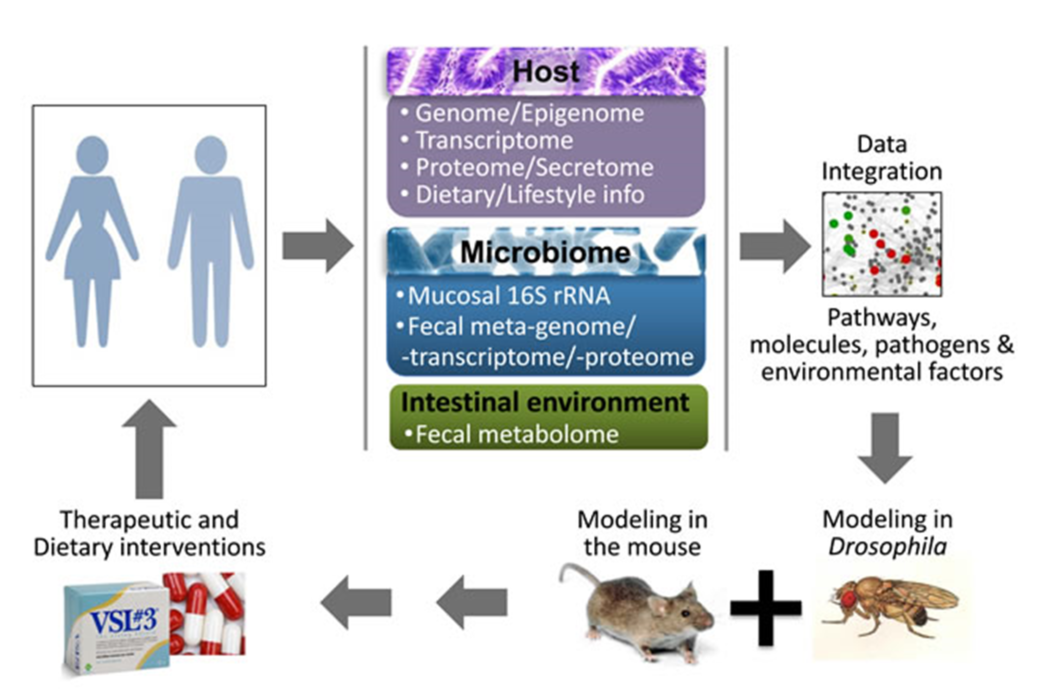
Figure: A roadmap to identify detrimental synergisms within human holo’omes as causal for colon cancer and develop personalized therapeutic or preventive strategies. A systems biology approach to assess shifts in intestinal holo’omes in humans and its link to colorectal pathologies will necessitate analysis of host intestine and microbial community genome, transcriptome, proteome metabolome and blood secretome. Using computational platforms, the genetic, metabolic, nutritional, microbial and immunological information accumulated, together with publicly available phenotypic and molecular function data, will be explored to obtain a ‘holistic’ view of key pathogenic processes and their hierarchies, to simulate the expected response to hypothetical interventions and develop new basic and translational research hypotheses. Reductionist approaches in Drosophila and mice - which can be genetically manipulated to express or lose the expression of specific genes in the intestine, while fed or injected with specific microbes and metabolites - could be used to assess detrimental synergisms of the intestinal holo’ome in driving inflammation and tumorigenesis, and guide the development of intervention strategies. Such therapeutic or dietary interventions could be translated to the clinic aiming to treat patients against microbial and intestinal environment imbalances as a means to alleviate intestinal inflammation and CRC.
